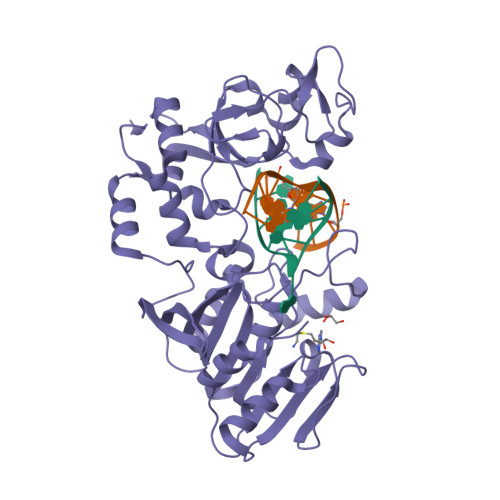
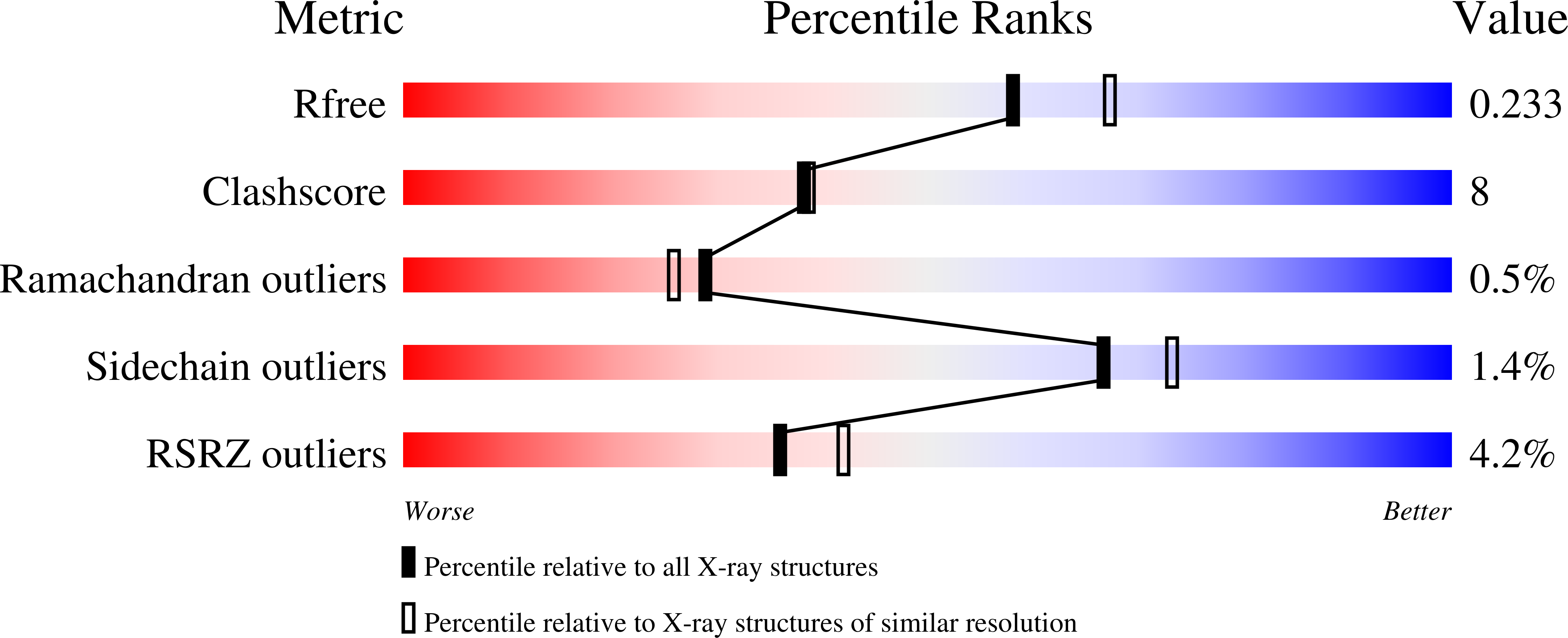
![RCSB PDB - 3O82: Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 5'-O-[N-(2,3-dihydroxybenzoyl)sulfamoyl] adenosine RCSB PDB - 3O82: Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 5'-O-[N-(2,3-dihydroxybenzoyl)sulfamoyl] adenosine](https://cdn.rcsb.org/images/structures/3o82_assembly-1.jpeg)
RCSB PDB - 3O82: Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 5'-O-[N-(2,3-dihydroxybenzoyl)sulfamoyl] adenosine
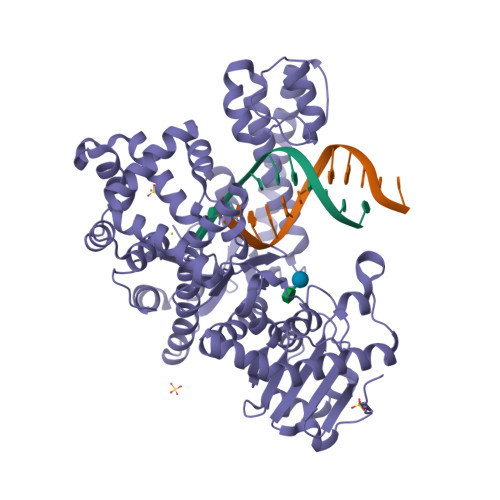
RCSB PDB - 1O15: THEOPHYLLINE-BINDING RNA IN COMPLEX WITH THEOPHYLLINE, NMR, REGULARIZED MEAN STRUCTURE, REFINEMENT WITH TORSION ANGLE AND BASE-BASE POSITIONAL DATABASE POTENTIALS AND DIPOLAR COUPLINGS

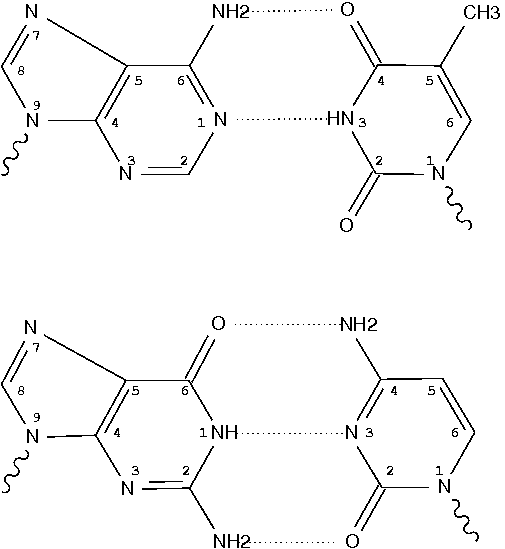
RCSB PDB - 150D: GUANINE.1,N6-ETHENOADENINE BASE-PAIRS IN THE CRYSTAL STRUCTURE OF D(CGCGAATT(EDA)GCG)

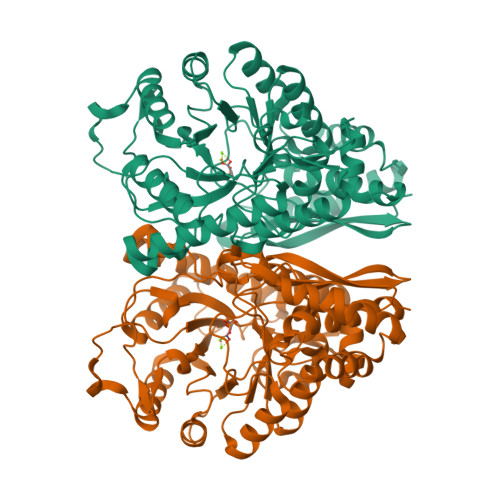
RCSB PDB - 2C7O: HhaI DNA methyltransferase complex with 13mer oligonucleotide containing 2-aminopurine adjacent to the target base (PCGC:GMGC) and SAH

RCSB PDB - 1YFL: T4Dam in Complex with Sinefungin and 16-mer Oligonucleotide Showing Semi-specific and Specific Contact and Flipped Base

RCSB PDB - 145D: Structure and thermodynamics of nonalternating C/G base pairs in Z-DNA: the 1.3 angstroms crystal structure of the asymmetric hexanucleotide D(M(5)CGGGM(5) CG)/D(M(5)CGCCM(5)CG)

RCSB PDB - 1C8C: CRYSTAL STRUCTURES OF THE CHROMOSOMAL PROTEINS SSO7D/SAC7D BOUND TO DNA CONTAINING T-G MISMATCHED BASE PAIRS

RCSB PDB - 2BR0: DNA Adduct Bypass Polymerization by Sulfolobus solfataricus Dpo4. Analysis and Crystal Structures of Multiple Base-Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine
RCSB PDB - 5DAR: CRYSTAL STRUCTURE OF THE BASE OF THE RIBOSOMAL P STALK FROM METHANOCOCCUS JANNASCHII
RCSB PDB - 255D: CRYSTAL STRUCTURE OF AN RNA DOUBLE HELIX INCORPORATING A TRACK OF NON-WATSON-CRICK BASE PAIRS

RCSB PDB - 1L3U: Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 11 base pairs of duplex DNA following addition of a dTTP and a dATP residue.
RCSB PDB - 6P0E: Human DNA Ligase 1 (E346A,E592A) bound to adenylated DNA containing an 8-oxo guanine:adenine base-pair

RCSB PDB - 1FUF: CRYSTAL STRUCTURE OF A 14BP RNA OLIGONUCLEOTIDE CONTAINING DOUBLE UU BULGES: A NOVEL INTRAMOLECULAR U*(AU) BASE TRIPLE